library(tidyverse)2 Functions
First, load the tidyverse.
2.1 Turning Procedures into Functions
A procedure is one or more lines of code we run to do things like calculate summary statistics, transform variables, and read and write files. Our existing code serves as raw material we can transform into functions, which makes it portable and allows us to reuse it within and across scripts with minimal repetition.
If you want to reuse a procedure more than two or three times, turn it into a function.
2.1.1 Example: Calculating the Standard Error
In the code below, we calculate the standard error (\(\frac{SD_x}{\sqrt{n}}\) ) of several columns in mtcars, which involves repeating the same procedure for several different R objects.
sd(mtcars$mpg) / sqrt(length(mtcars$mpg))[1] 1.065424sd(mtcars$wt) / sqrt(length(mtcars$wt))[1] 0.1729685sd(mtcars$hp) / sqrt(length(mtcars$hp))[1] 12.12032The functions are the same, but the objects vary.
Two things change in each line: the object inside of sd() and the object inside of sqrt(length()), which are the same object within each line. Fill them in with a placeholder, x:
sd(x) / sqrt(length(x))Now, to turn this procedure into a function, we need (1) a name and (2) the number of arguments, with suitable names.
For the name of the function, se is concise and meaningful, but we should first check if it is the name of another existing function in a library we have loaded at the moment:
?seNo documentation for 'se' in specified packages and libraries:
you could try '??se'That means it is not already used, so we will not have any naming conflicts if we were to use this name.
For the arguments, we only have one argument, which we can keep calling x. If we had multiple arguments or planned on using a function more often, we should give the arguments more informative names. For now, x is fine.
The structure of our function-making syntax is as follows:
function_name <- function(argument) {
procedure
}If we replace function_name, argument, and procedure with their values, we get this code:
se <- function(x) {
sd(x) / sqrt(length(x))
}Running that line will add se() to our global environment under the Functions header, so that is available for our use. Now let’s use it!
se(mtcars$mpg)[1] 1.065424se(mtcars$wt)[1] 0.1729685se(mtcars$hp)[1] 12.120322.1.2 Example: Counting Missing Values
As another example, let’s get the number of missing values in a vector:
sum(is.na(airquality$Ozone))[1] 37sum(is.na(airquality$Solar.R))[1] 7What changes in each line? Replace that with x:
sum(is.na(x))Name the function nmiss and fill in the function template:
nmiss <- function(x) {
sum(is.na(x))
}Now use it:
nmiss(airquality$Ozone)[1] 37nmiss(airquality$Solar.R)[1] 72.2 Exercises: Basic Functions
The code below returns the proportion of rows in a dataframe that do not have any missing data. Create a function called
prop_observed()that does the same.nrow(na.omit(txhousing)) / nrow(txhousing)[1] 0.828412nrow(na.omit(storms)) / nrow(storms)[1] 0.1110713prop_observed(txhousing)[1] 0.828412prop_observed(storms)[1] 0.1110713The following line returns how many years have passed since some date given in year-month-day format. In this example, we use July 20, 1969, the Apollo 11 moon landing.
time_length(interval(ymd(19690720), Sys.Date()), unit = "years")[1] 55.70411Create a function called
years_since()that does the same, and use it to find how many years have passed since some significant dates for you, like a birthday, wedding, or starting your graduate school. (Note theymd()function works with dates in various year-month-day formats, everything from19690720to"1969-07-20"to"1969 July 20". For working with dates, see our chapter on date vectors.)years_since(19690720)[1] 55.70411The code below prints a greeting.
cat(paste0("Good morning, ", "Jason", ".\nToday is ", format(Sys.Date(), "%B %d, %Y"), ".\n"))Good morning, Jason. Today is April 03, 2025.Create a function called
morning()that takes the user’s name.morning("World")Good morning, World. Today is April 03, 2025.
2.3 Environments
Most of our work in R involves working with objects in the global environment. This is the one we see in the top-right Environment pane of RStudio, and the objects there are available to everything in our current session. Other environments exist, and functions actually execute in their own environment.
The technical details of environments are discussed elsewhere1, so here we will focus on the implications for our function writing. Keep these three things in mind:
- Objects created within functions with
<-are not stored in the global environment. - Each time a function is run, it runs in a new environment.
- A function searches its own environment before it searches the global environment.
We will discuss each of these in turn.
2.3.1 Objects created within functions
Let’s create a somewhat useful function that reports the empirical standard deviation from a random sample drawn from a normal distribution of size n. The expected standard deviation, per the default of rnorm(), is 1. The error tends to get smaller as the sample size increases.
empirical_sd <- function(n) {
sample_data <- rnorm(n)
sd(sample_data)
}
empirical_sd(10)[1] 0.780586empirical_sd(10000)[1] 1.012406empirical_sd(1000000)[1] 1.000062Take a look at the procedure inside the function. We make an object, sample_data, and then calculate and print the standard deviation.
Does sample_data exist in our global environment?
"sample_data" %in% ls()[1] FALSENo. It only exists within the function’s environment. Functions do not return intermediary assigned objects; instead, they return the result of the last expression.
2.3.2 Functions run in new environments
Not only do functions run in their own environments, they run in a new environment each time.
The function add_one() has no arguments, and it simply adds one to the object i. We initialize it with a value of 1. After the first call of add_one(), i is 2. We may expect that i increases each time, but it does not.
add_one <- function() {
i <- i + 1
i
}
i <- 1
add_one()[1] 2add_one()[1] 2add_one()[1] 2The reason for this is that our global environment has i with a value of 1. The first call of add_one() creates an environment, gets i from the global environment, and adds 1 to it, so that i is now 2. The next call of add_one() does not see the environment from the previous call of the function, so it gets i with a value of 1 from the global environment, adds 1 to make 2, and so on.
The function will search its own environment before searching the global environment. The upside of this is that we do not need to worry about conflicts between function argument names and our global environment.
2.3.3 Environment Search Order
To illustrate the search order of functions, let’s modify the add_one() code from above to include the line i <- 10. The output is 11 because add_one() finds the i with a value of 10 in its own environment before finding the i with a value of 1 in the global environment.
add_one <- function() {
i <- 10
i <- i + 1
i
}
i <- 1
add_one()[1] 11The upside of this is that we do not need to worry about conflicts between the names of objects in our function, and the names of objects in our global environment. That is, while in general we need to thoughtfully name our objects, we are also free inside our functions to call every vector x or y and every dataframe dat or df.
2.4 Multiple Returns
Given that assigning objects within functions does not assign objects in our global environment, and our last returned object is returned, how can we return multiple objects?
Let’s add to nmiss() to also give the total number of values and the proportion of missing values:
miss_stats <- function(x) {
length(x)
sum(is.na(x))
mean(is.na(x))
}miss_stats(airquality$Ozone)[1] 0.2418301Only the last value is returned, mean(is.na(x)).
A solution is to make a (named) list that contains all the objects we want returned. Lists are excellent containers when you want to return a variety of objects since they can contain objects of different types and structures.
miss_stats <- function(x) {
list(n = length(x),
miss_count = sum(is.na(x)),
miss_prop = mean(is.na(x)))
}miss_stats(airquality$Ozone)$n
[1] 153
$miss_count
[1] 37
$miss_prop
[1] 0.24183012.5 Arguments and Defaults
If we use more informative argument names, our functions become easier to use and later understand.
The code below takes proportions (0-1) and turns them into percentages (0-100%) with a differing numbers of decimal places.
x <- seq(0, 1, .1243)
y <- seq(0, 1, .2345)x[1] 0.0000 0.1243 0.2486 0.3729 0.4972 0.6215 0.7458 0.8701 0.9944y[1] 0.0000 0.2345 0.4690 0.7035 0.9380trimws(paste0(format(round(x * 100, 2), nsmall = 2), "%"))[1] "0.00%" "12.43%" "24.86%" "37.29%" "49.72%" "62.15%" "74.58%" "87.01%"
[9] "99.44%"trimws(paste0(format(round(y * 100, 1), nsmall = 1), "%"))[1] "0.0%" "23.4%" "46.9%" "70.3%" "93.8%"In these, we have two pieces that change: the vector (x, y) and the number of decimal places (1, 2), which occures twice. We can use x for the vector argument name and n_decimal for the number of decimal places argument name.
trimws(paste0(format(round(x * 100, n_decimal), nsmall = n_decimal), "%"))The first line of our code to create that function will look like this:
prop_to_perc <- function(x, n_decimal) {We may find that we often want to round to two decimal places, so we should set that as an argument default. Defaults are set with the pattern argument = value:
prop_to_perc <- function(x, n_decimal = 2) {Now, reformat the code inside the function with pipes to make it more readable.
prop_to_perc <- function(x, n_decimal = 2) {
round(x * 100, n_decimal) |>
format(nsmall = n_decimal) |>
paste0("%") |>
trimws()
}Now use the function on x and y. For x, we are fine with the default of 2 decimal places, so we do not need to specify anything.
prop_to_perc(x)[1] "0.00%" "12.43%" "24.86%" "37.29%" "49.72%" "62.15%" "74.58%" "87.01%"
[9] "99.44%"And for y, we want just 1 decimal place, so we need to specify that:
prop_to_perc(y, n_decimal = 1)[1] "0.0%" "23.4%" "46.9%" "70.3%" "93.8%"Following our earlier discussion of environments, note how each function call finds the x in its own environment instead of the x in the global environment.
2.6 Conditionals
Sometimes we may want a single function that does different things with different types of input.
For example, take the function plot(). If we give it a numeric vector, it gives us a scatterplot of values by index:
plot(chickwts$weight)
And if we give it a factor, it returns a bar chart:
plot(chickwts$feed)
But it fails if we give it a character vector:
plot(as.character(chickwts$feed))Warning in xy.coords(x, y, xlabel, ylabel, log): NAs introduced by coercionWarning in min(x): no non-missing arguments to min; returning InfWarning in max(x): no non-missing arguments to max; returning -Inf
Error in plot.window(...): need finite 'ylim' valuesAnd if we give it a logical vector, it returns a plot where TRUE and FALSE have been coerced to 1 and 0, respectively:
plot(chickwts$weight > 250)
Let’s make a function that plots vectors of different types. It can return the default plots for numeric and factor vectors, but for character and logical vectors, it should coerce them to factor vectors first.
We should also limit the function to only work with vectors so that we do not get unexpected results with other data structures and objects. To do this, we can use stopifnot() with a condition: check if it is a vector or a factor (which technically is not a vector…):
stopifnot(is.vector(x) | is.factor(x))After that, we need branching logic. If it is a character or logical vector, make it a factor and then plot it. Otherwise, plot it as-is:
if (is.character(x) | is.logical(x)) {
plot(as.factor(x))
} else {
plot(x)
}Put this all together inside a function which we can call plot_vec():
plot_vec <- function(x) {
stopifnot(is.vector(x) | is.factor(x))
if (is.character(x) | is.logical(x)) {
plot(as.factor(x))
} else {
plot(x)
}
}And now test it with a few inputs:
plot_vec(chickwts$weight) # numeric
plot_vec(chickwts$feed) # factor
plot_vec(as.character(chickwts$feed)) # character
plot_vec(chickwts$weight > 250) # logical
Now give it another data structure to see how it reacts:
plot_vec(mtcars)Error in plot_vec(mtcars): is.vector(x) | is.factor(x) is not TRUEIt is working as expected, but we could improve that error message.
We can change from using stopifnot() to using an if statement with our condition along with stop(), which can contain a more helpful error message. It can tell the user the structure of the provided object and instruct them to instead supply a vector.
plot_vec <- function(x) {
if (!(is.vector(x) | is.factor(x))) {
stop(paste("x is a", class(x)[[1]], "but it should be a vector"))
}
if (is.character(x) | is.logical(x)) {
plot(as.factor(x))
} else {
plot(x)
}
}Now try it out:
plot_vec(mtcars)Error in plot_vec(mtcars): x is a data.frame but it should be a vector2.7 Tidyverse Functions
tidyverse functions usually make code easier to read and write because we do not need to repeat the dataframe name each time2:
df$var3 <-
df$var1 + df$var2
df <-
df |>
mutate(var3 = var1 + var2)In the context of functions, however, using tidyverse functions requires extra work since we need to explicitly tell them to look within the dataframe to find the variables.
To illustrate the problem, here we make a function called wrong() that tries to get the mean of a given variable, but it fails to find mpg since it does not know to look inside mtcars. Adding quotes around the variable name does not solve the issue either.
wrong <- function(df, var) {
df |> summarize(mean = mean(var))
}
wrong(mtcars, mpg)Warning: There was 1 warning in `summarize()`.
ℹ In argument: `mean = mean(var)`.
Caused by warning in `mean.default()`:
! argument is not numeric or logical: returning NA mean
1 NAwrong(mtcars, "mpg")Warning: There was 1 warning in `summarize()`.
ℹ In argument: `mean = mean(var)`.
Caused by warning in `mean.default()`:
! argument is not numeric or logical: returning NA mean
1 NAThis issue can be solved in one of two ways within our function:
- Embrace the variable name with double curly braces:
{{ var }}. Seeright1()below. - Use the
.datapronoun with a character vector:.data[[var]]. Seeright2()below.
right1 <- function(df, var) {
df |> summarize(mean = mean({{ var }}))
}
right1(mtcars, mpg) mean
1 20.09062right2 <- function(df, var) {
df |> summarize(mean = mean(.data[[var]]))
}
right2(mtcars, "mpg") mean
1 20.09062When would we use one over the other?
{{ var }} is useful when we specify variable names in function arguments
.data[[var]] is useful when we have a character vector returned by something else, like user input from a Shiny app or output from some other code like
myVar <- colnames(mtcars)[[1]]
myVar[1] "mpg"2.7.1 Embracing
We often need to calculate custom summary statistics for variables in our data. The code below takes a dataframe and variable as arguments, so we need to embrace the variable name in the function syntax:
mySummary5 <- function(df, var) {
df |>
summarize(n = n(),
mean = mean({{ var }}, na.rm = T),
median = median({{ var }}, na.rm = T),
sd = sd({{ var }}, na.rm = T),
se = sd({{ var }}, na.rm = T) / sqrt(n()))
}
mySummary5(airquality, Temp) n mean median sd se
1 153 77.88235 79 9.46527 0.7652217mySummary5(airquality, Ozone) n mean median sd se
1 153 42.12931 31.5 32.98788 2.666912(Note that we could have used .data[[var]] within the function and then specified “Temp” and “Ozone” in the function calls.)
2.7.2 The .data Pronoun
In this example, let’s fit a regression model and then produce plots to assess the linearity assumption. Because we want to specify the variable names as character vectors, we need to specify .data[[var]] within the function.
mod <- lm(mpg ~ wt + disp + drat, mtcars)
linearity_plot <- function(df, mod, var) {
df <-
df |>
mutate(res = residuals(mod),
yhat = fitted(mod))
ggplot(df, aes(.data[[var]], res)) +
geom_point() +
geom_hline(yintercept = 0, color = "red") +
geom_smooth()
}Now use the function to plot the fitted values and each predictor (just wt here, but we should check all of them):
linearity_plot(mtcars, mod, "yhat")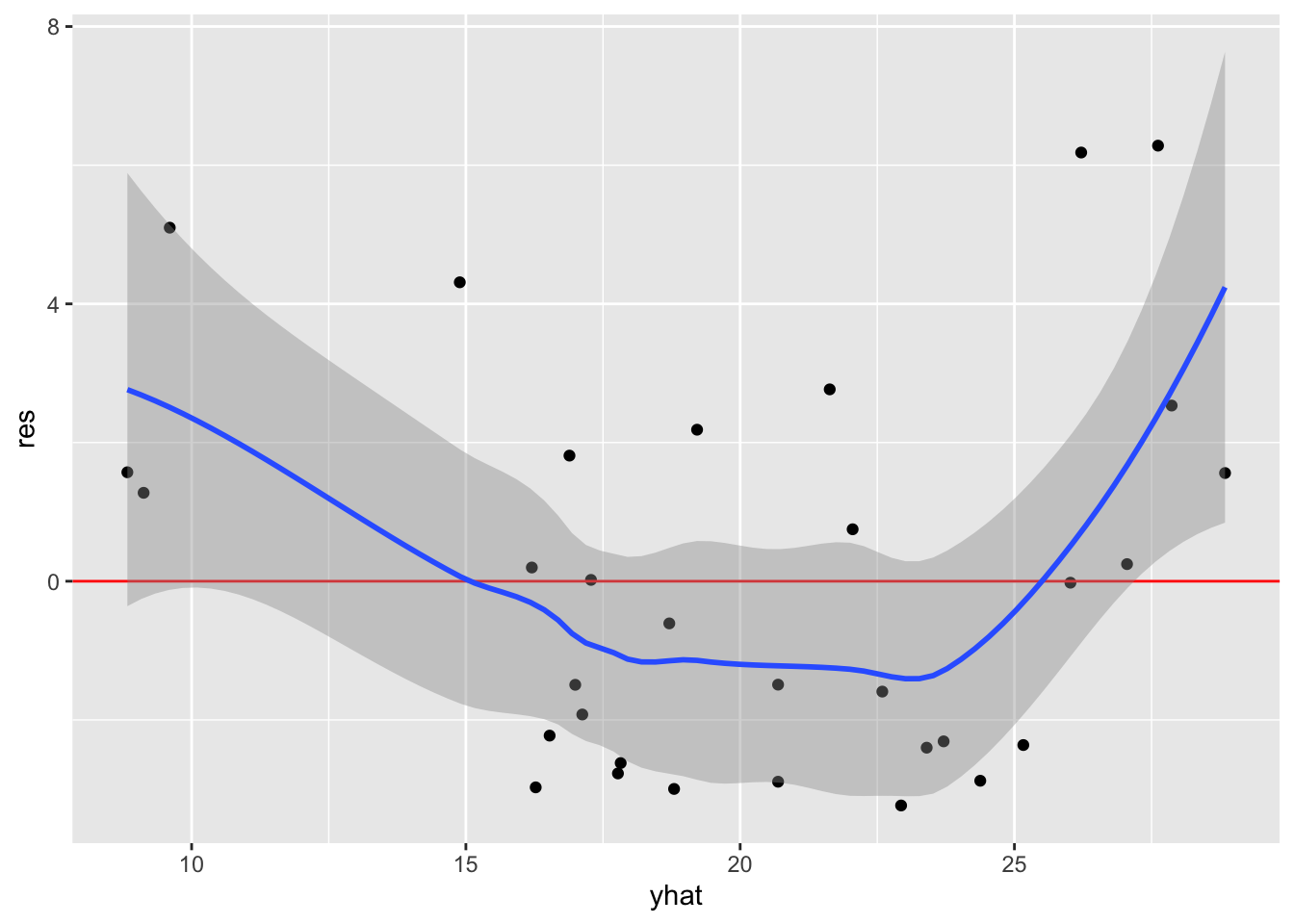
linearity_plot(mtcars, mod, "wt")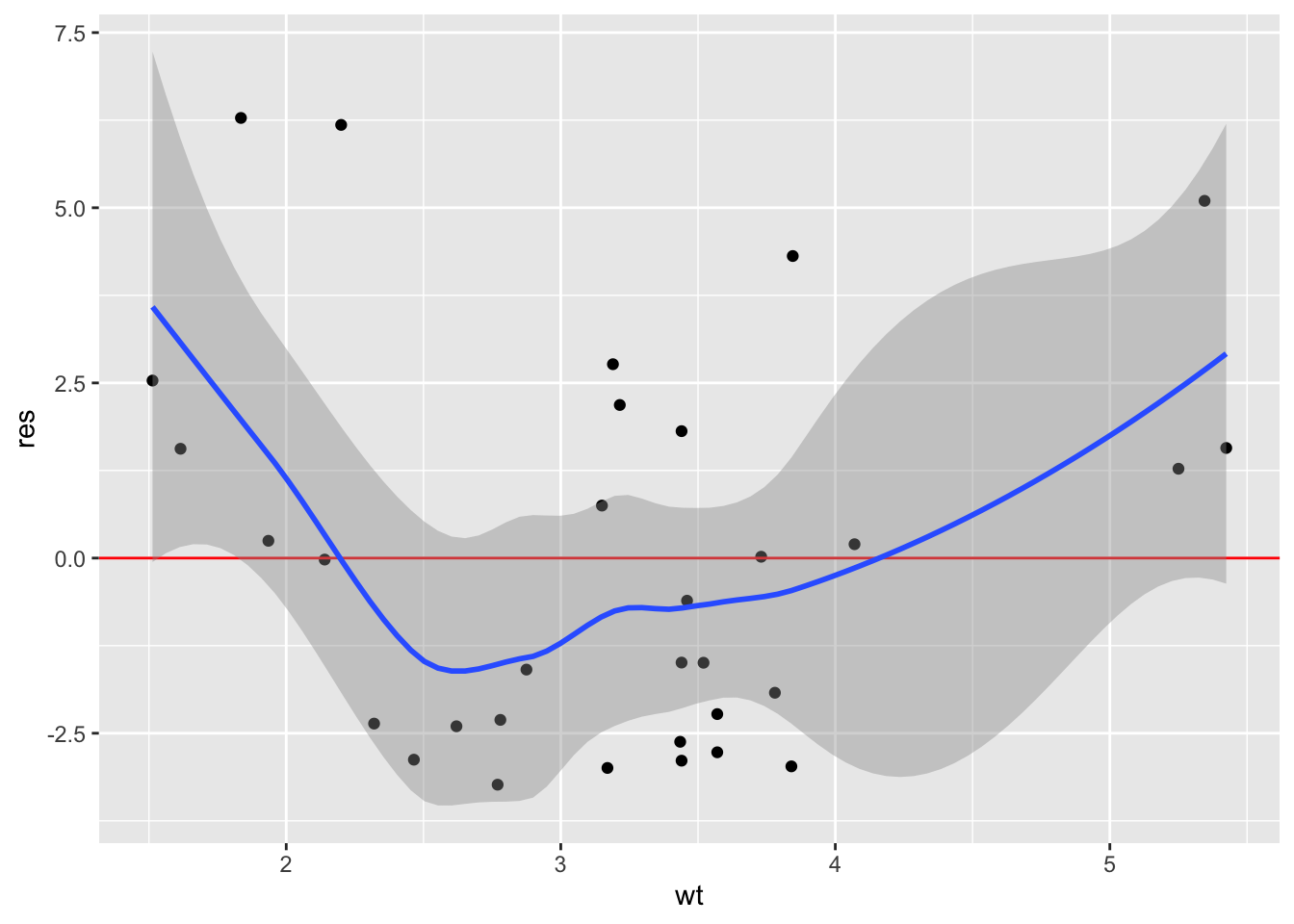
(Note we could have used embracing ({{ var }}) in the function and then referred to the vars as yhat and wt in the function calls.)
To show where we will go in the next chapter on Iteration, we can first programmatically extract the names of our predictors and then iteratively plot them with map():
predictors <- colnames(model.frame(mod))[-1]
predictors[1] "wt" "disp" "drat"map(predictors, \(x) linearity_plot(mtcars, mod, x))[[1]]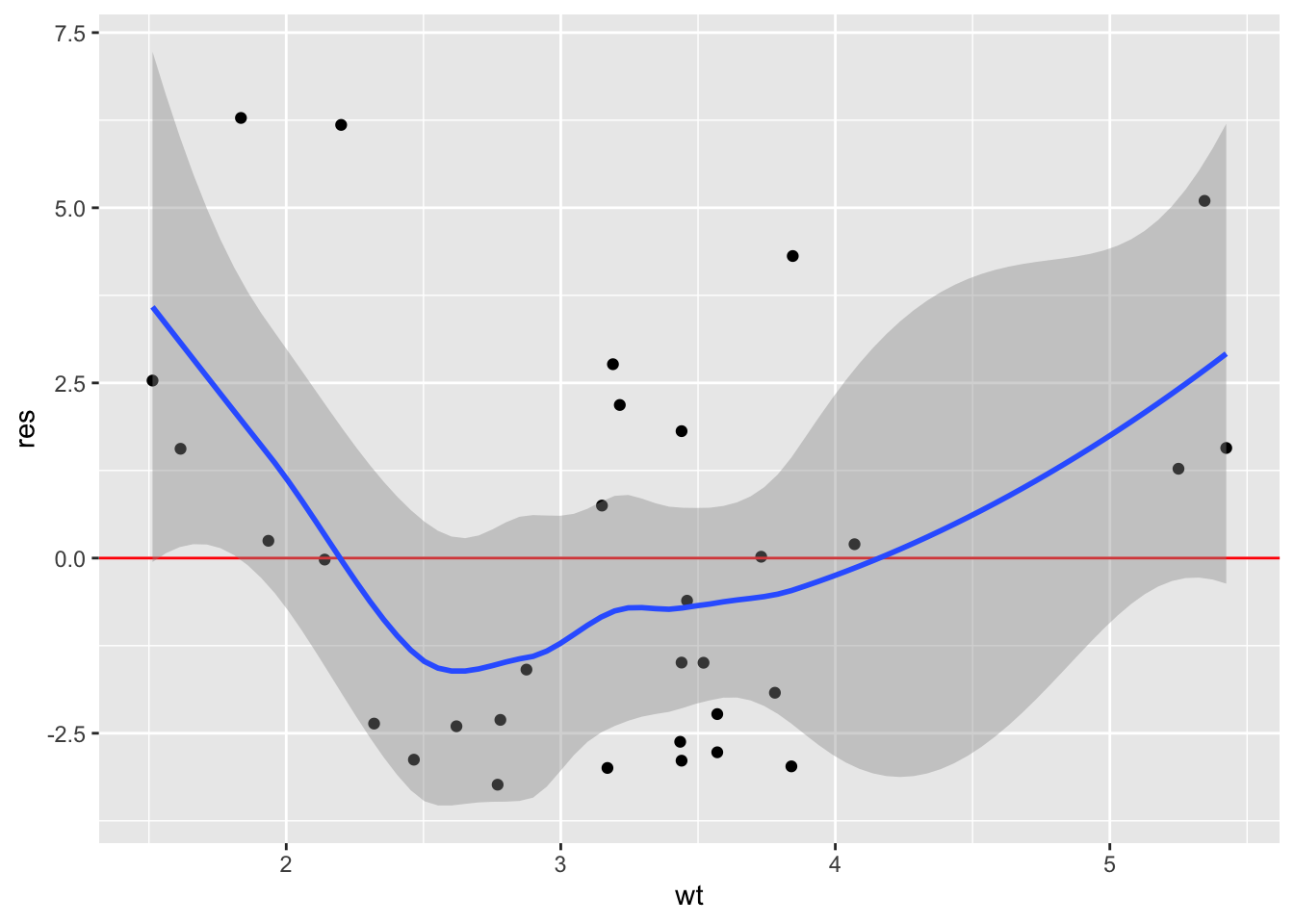
[[2]]
[[3]]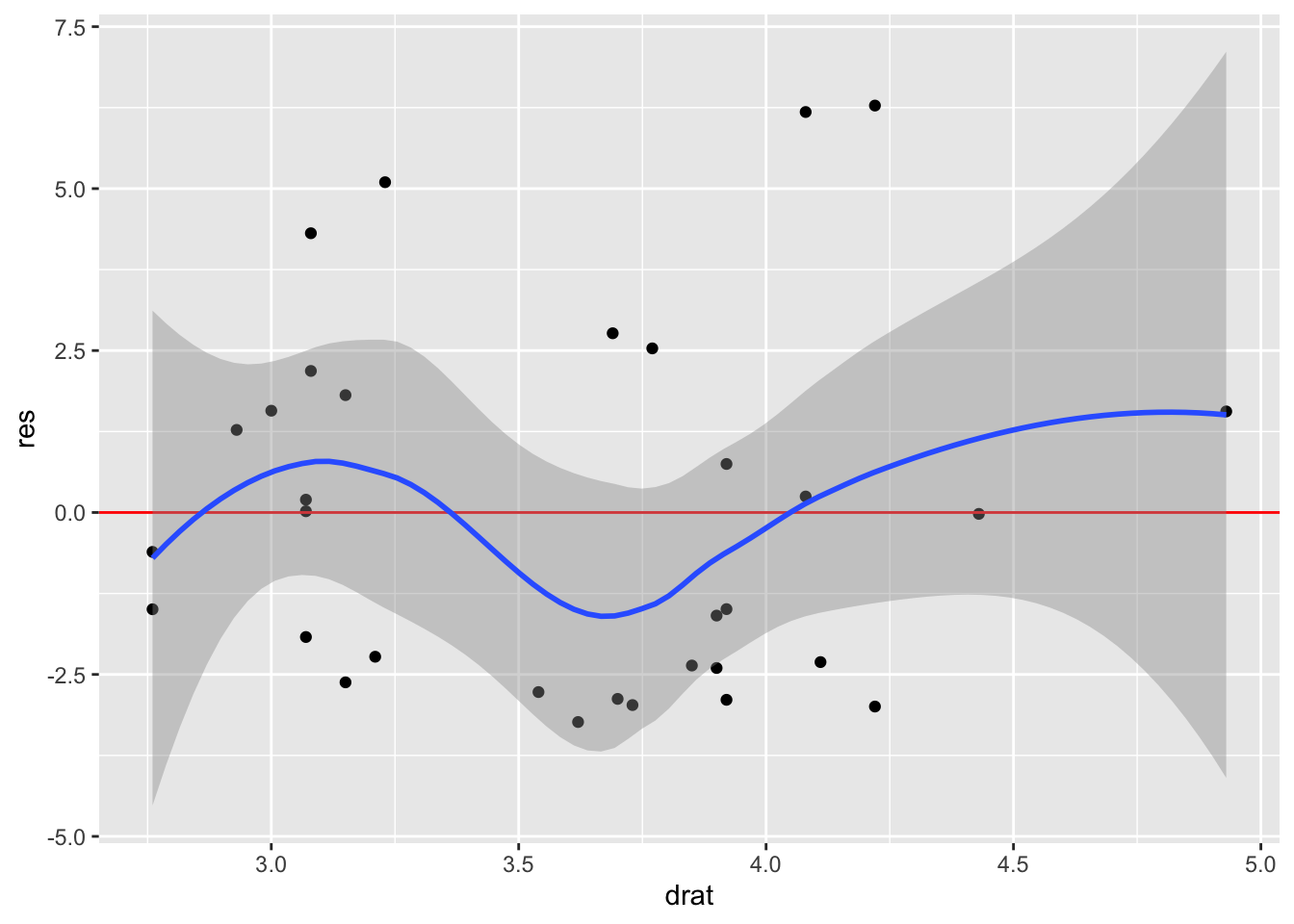
2.8 Reusing Functions Across Scripts
We may find that some functions we write are actually quite useful, and we want to use them across projects. We know we should not copy and paste code again and again within a script, but what about between scripts? How can we make a function available to multiple projects?
We have two options: creating a script with functions, and writing a package. Package writing is outside the scope of this resource. Packages can be used locally without publishing them, but it would be quite an addition to your resume to publish packages to GitHub or even CRAN. See R Packages to learn more.
For now, let’s just put our functions inside a script somewhere on our computer. We then need to “source” that script. Just add a line to the beginning of your script where you load other libraries:
library(tidyverse)
source("path/to/file/myFunctions.R")source() will run a script. Any functions or objects created in that script will be added to the global environment, and then you can use them in your current project.
2.9 Exercises: Complex Functions
Revisit the time difference calculator from the previous exercises. Modify the function so that the units (years, months, days, etc.) can be specified.
time_since(19690720, "days")[1] 20346Sometimes, missing is coded with more than one missing code. Write a function that uses
ifelse()and returns a vector after replacing specified values with NA. For example, if 8 and 9 are missing codes, this vector:set.seed(2) x <- sample(c(1:3, 8, 9), 10, replace = T) x[1] 9 1 9 1 8 9 1 2 3 1should be returned as:
to_na(x, 8:9)[1] NA 1 NA 1 NA NA 1 2 3 1Fix this function. The problem is that the output does not vary.
get_sample()should return a sample of size n. Fix it so thatget_sample(3)returns 3 values. Refer to?sample.get_sample <- function(n) { sample(1:5, replace = T) } get_sample(3)[1] 3 2 3 1 1Fix this function. The problem is that there is no output.
fit_mod()should return a fitted model object, but nothing is returned.fit_mod <- function(df, outcome, predictors) { form <- as.formula( paste(outcome, "~", paste(predictors, collapse = "+") ) ) mod <- lm(form, data = df) } fit_mod(df = mtcars, outcome = "mpg", predictors = c("wt", "disp", "wt*disp"))Fix this function. The problem is that the scatterplot only shows one point.
scatter()should produce a scatterplot of a givenxandyfrom dataframedf.scatter <- function(df, x, y) { ggplot(df, aes(x, y)) + geom_point() } scatter(mtcars, "wt", "mpg")
To learn more about environments in R, see the “Environments” chapters of Advanced R and Hands-On Programming with R.↩︎